Recently, scientists at the Stanford group in association with the J. Craig Venter Institute published a paper regarding the successful attempt to simulate a complete organism from its DNA in a computer cluster, for the first time in the history. They simulated a simplest bacterium named Mycoplasma Genitallium, which is capable of replicating inside the simulation. The implication of this is enormous, as this will bridge the gap between engineering and biology and in our quest for solutions to many problems. You can read about the analysis in Nature.com here. The original paper here
Seven months back, on my sabbatical, I was trying to do the same.
I was trying to simulate a simple bacterium from DNA available public from here. It has only one circular chromosome. I learned about the processes involved in the cell formation from the DNA. DNA is basically the code for two fundamental things happening inside an organism – replication and transcription. Replication is the process of making a copy of the DNA itself and transcription is the process of reading the gene code blocks and generating long chain molecules called Proteins in multiple steps.
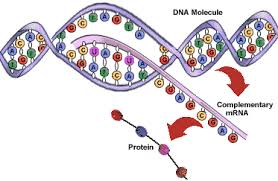
DNA transcription
My work is more inspired by the theory of cellular automata and the Conway’s game of Life. I also considered the concept of fractal to model my organism.
In order to understand the DNA we have to look at it through an evolutionary perspective. If you look at a particular organism or more specifically its genotype and its phenotype, its just a small snapshot of an unbroken chain of repetitive process of replication(the output is a copy of the DNA) and transcription(the output is proteins ). The process started since the beginning of life itself and have undergone innumerable changes(evolution). This in my opinion is a fractal pattern.
Mathematicians have shown before that simple life like structures can be represented by simple set of equation iterated numerous times. A fern for eg. Barnsley fern.

Barnsley Fractal Fern
Instead of placing dots in an image as done by the Barnsley fern equation, the DNA puts complex 3D structures with lot of different physical properties called proteins. This is similar but complex form of Conway’s Game of Life, but much complex. In Game of Life, the cells are actually square cells which can have only one of two states : “dead” or “alive”. From simple rules like ” if a cell is surrounded by at most three living cell, the cell will replicate else it will die”, we can see self sustaining life like activities emerge from the computer screen from a set of original points.

Bill Gosper’s Glider Gun in action—a variation of Conway’s Game of Life. This image was made by using Life32 v2.15 beta, by Johan G. Bontes. (Photo credit: Wikipedia)
The DNA is the simple set of equations that give raise to complex structures if allowed to repeat enormous time. The structure (phenotype) we see is an emerged form from this process of protein making and other associated process which are perceived as phenotype. For eg. snow flakes are beautiful complex patterns emerge from the simple physical laws.

Snow Flakes – Fractal like structures emerging from natural processes. Example of emergence.
The hypothesis is that if we can model basic blocks of the DNA, which are the Carbon, Hydrogen, Oxygen and Nitrogen atoms, and simulate all the dynamics between them (the fractal equation) and create conditions similar to a culture medium, the living bacteria will emerge from it! If we can put ultra-fast computers to perform probabilistic calculations and molecular level interaction.
Of-course it will be very difficult for one amateur guy like me to attempt, but I want to abstract many levels and assume certain things taken for granted.
In contrast with the simulation of whole cell by simulating all smaller sub units, which is a top-down approach, my approach is bottom up – simulate the lowest building block accurately and iterate interactions as it will naturally occur based on probability, predicting that life like features will emerge.
I am on sabbatical again and hope to make it happen this time. Visit this blog later to see if I am successful. Or tweet to me @anandcv
